Recently a research paper titled “Expanding the Diversity of Mycobacteriophages: Insights into Genome Architecture and Evolution” was published in PLoS ONE, a peer-reviewed online journal published by the Public Library of Science.
The authors included 12 Washington University undergraduates who had participated as freshmen in the inaugural Phage Hunters course at Washington University in St. Louis.
Phages are viruses that infect bacteria by injecting genetic material into them with a syringe-like plunger. In fact, they even look rather like outlandish syringes.
Phage Hunting
Phage Hunters is a freshman focus course the biology department first offered in 2008. The course, which is supported by the Science Education Alliance at the Howard Hughes Medical Institute, offers freshmen a research experience isolating and characterizing novel phages.
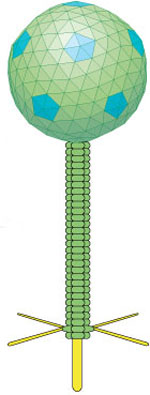
ViralZone 2010, Swiss Institute of Bioinformatics
The St. Louis phages belong to a group called Siphoviridae that have non-contractile tails and icosahedral heads. (The name comes from the Greek word sipho, meaning tube and refers to the long tail.) The phage uses its tail fibers to attach to a target cell and then breaks down the cell wall before injecting its DNA into the host. The viral DNA than co-opts the target cells cellular machinery to make more phages, which eventually burst out of the cell.
Starting in the fall semester, students collect soil samples, and isolate phages from the soil. Once isolated, students purify and characterize the phage. Nearly all the phages isolated in this way are new to science.
Over winter break the DNA genomes of several phages are sequenced, and in the spring semester students analyze the sequence information to get an in-depth picture of how their phage works and relates genetically to other previously characterized phages.
“A critical part of science education is helping students learn to ‘think like a scientist,’ says Sarah C. Elgin, PhD, the Viktor Hamburger Distinguished Professor in Arts & Sciences, who taught the class. “This course requires students to do just that, giving them new analytical skills and a new perspective. Plus, they have a lot of ownership of the project — it is their phage — something that no one else has studied. Discovery is a real thrill!”
More on Phages
Phage are the most numerous “biological entities” in the world (they have to be called entities because no one is quite sure whether they are alive in any practical sense).
There are thought to be 1031 virus particles floating about, and 1023 phage infections a second.
Most specialize in infecting a particular species of bacteria. For the class the host bacterium was Mycobacterium smegmatis, which is related to the bacteria that cause leprosy and tuberculosis, but does not itself cause disease.
Because it is a fast grower, M. smegmatis has become a laboratory workhorse.
About 70 phages that prey on mycobacteria have been previously described. The authors of the PloS ONE paper, who were then freshmen at 12 colleges and universities, reported the isolation, sequencing, and annotation (labeling parts of the genome whose function is known) of 18 new ones.
Angelica and Uncle Howie
The new mycobacteriaphages included two found by the WUSTL students. One, named Angelica, was isolated from soil in Clayton, Mo. The other, called Uncle Howie, was isolated from soil in University City, Mo. Both Clayton and University City are suburbs of St. Louis.
(The students have “naming rights” for the phage they isolate. Consequently, other phage also have amusing names, including Corndog, Fruitloop, Tweety, Predator and Gumball. )
Most of the new phages, including the two St. Louis ones, had what is called siphoviral morphologies — long flexible non-contractile tails, and isometric icosahedral heads.
Once the students had sequenced the phages, they set out to compare their genomes to those of other phages. Bacteriophage genomes are mosaics or assemblages of modules, each of which corresponds to a gene or group of genes.
Since the phages are thought to have evolved by the exchange of these modules, comparing phages can provide clues to their evolutionary history.
The mycobacteriophages seem to fall into nine major clusters, with a few leftovers, or singletons, with no close relatives. Angelica, the Clayton phage, fell into cluster K and Uncle Howie, the University City phage, in cluster B.
There has been considerable speculation about the mechanisms that give rise to the mosaic genomes and the work also provided some insight into this problem. The clusters of phages indicate lateral transfer of genes or DNA fragments from one phage to another, as well as inheritance by descent.
The WUSTL student authors are: Samantha T. Alford, Alexander G. Anderson, William D. Barshop, Lisa Deng, Vincent J. Huang, Peter M. Hynes, Patrick C. Ng, Hannah Rabinowitz, Corwin Rhyan, Erika F. Sims, Emilie G. Weisser, and Bo Zhang.
In addition, the graduate student teaching assistant, Anthony Tubbs, and instructors Elgin, Kathy Hafer, PhD, senior lecturer in biology, and Chris Shaffer, research associate in biology, are co-authors on the paper.
“It is very rewarding to see students so engaged in the work we do in the Phage Hunters course,” Hafer says. “Many really blossom as they gain confidence in their biology knowledge and research ability. And publication validates their membership in the science community.”